Windows Installation¶
In this section we describe how to install all the necessary requirements and set up the Clinical Knowledge Graph on a Windows operating system.
Neo4j¶
The installation of Neo4j on Windows follows the same steps as in Installing Neo4j
Python¶
The installation of Python on Windows follows the same steps as in Installation
Microsoft Visual C++ Build Tools¶
Running python on Windows can sometimes result in the following error:
error: Microsoft Visual C++ 14.0 or greater is required.
To fix this error, you will need to download and install Microsoft Build Tools for Visual Studio.
Once installed, click Workloads, select all the packages available and install them. This will require several Gigabytes of disk space so, as an alternative and if your machine has limited space, you can install only C++ build tools under Workloads, and Windows 10 SDK and the latest version of MSVC v142 - VS 2019 C++ x64/x86 build tools under Individual Components.
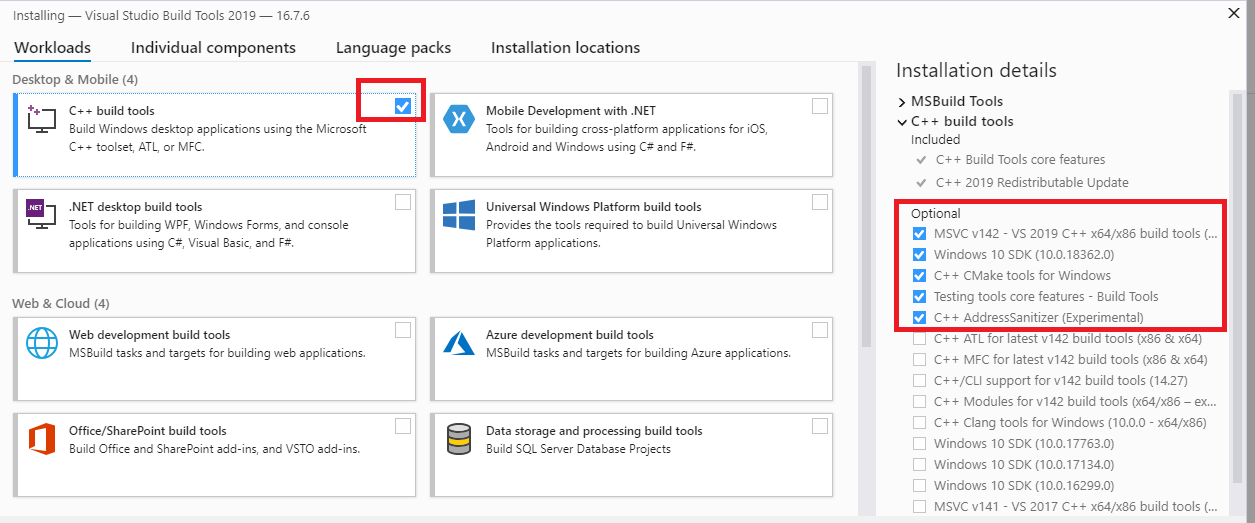
The build tools allow Python packages to be built in Windows, from the command line (MSVC cl.exe module is used as a C/C++ compiler).
R¶
Another essential package for the functioning of the Clinical Knowledge Graph is R. R packages (SAMR and WGCNA) are not available for Windows yet due to library incompatibilities (Rpy2). We are however working to solve this to provide the full functionality also in Windows.
You can check if an R version >= 3.6.1 is already installed by running:
> r --version
If R is not installed in your machine, please follow these tutorial.
In order to simplify calling R from the command prompt, you can choose to add it to PATH and to the environment variables. To do so, follow the steps bellow:
Go to the Windows menu, right-click on Computer and click on Properties.
From the computer properties dialog, select
Advanced system settingson the left panel. And from there, click on Environment variables button.In the Environment variables dialog, click the New button in the top half of the dialog, to make a new user variable.
Give the variable name as
Rand the value is the path to the R executable, which is usuallyC:\Program Files\R\R-4.0.0\bin\R.exe.In the bottom half of the Environment variables dialog, find the variable
Path, select it and click Edit.In the edit dialog window, add
;to the end of the variable value followed by the R path used when creating the previous environmental variable.Click OK to save, click OK and OK again to save the new variable and edit to
Path.
To run R from the command line, run:
> R
All R packages can be installed by simply initiating R (command prompt or R shell) and running:
install.packages('BiocManager')
BiocManager::install()
BiocManager::install(c('AnnotationDbi', 'GO.db', 'preprocessCore', 'impute'))
install.packages(c('devtools', 'tidyverse', 'flashClust', 'WGCNA', 'samr'),
dependencies=TRUE, repos='http://cran.rstudio.com/')
install.packages('IRkernel')
Warning
If the install does not work (cannot write to library), run a new command prompt as administrator:
Go to the Windows menu, right-click on Command Prompt and select
Run as administrator.
In this new prompt, launch R and run the previous R install packages.